來源: https://www.kaggle.com/corochann/covid-19-current-situation-on-december/notebook
此筆記的資料集來源: Johns Hopkins University github repository
最後更新: 2021-02-01
主要重點
- 綜觀全球疫情走勢
- 以
折線圖分析
- 以
- 分析各國確診與死亡人數變化
- 以
柱狀圖、顏色指標全球動態地圖、顏色指標折線圖、顏色指標熱圖分析
- 以
- 各國省分詳細資訊
- 有多少國家提供精確的省分疫情資料
- 以
Dictionary分析
- 深入探討美國疫情
- 以
表格、顏色指標美國動態地圖、顏色指標折線圖、顏色指標熱圖分析
- 以
- 分析歐洲疫情
- 以
顏色指標歐洲動態地圖、顏色指標折線圖、顏色指標熱圖分析
- 以
- 分析亞洲疫情
- 以
顏色指標亞洲動態地圖、顏色指標折線圖、顏色指標熱圖分析
- 以
- 哪些國家已經脫離疫情高峰
- 以
顏色指標全球動態地圖分析
- 以
- 全球疫情何時趨緩
- 使用 sigmoid 函數預測
- 以
顏色指標折線圖分析
安裝
pip install -r requirements.txt
import gc
import os
from pathlib import Path
import random
import sys
from tqdm import tqdm
import numpy as np
import pandas as pd
import scipy as sp
import matplotlib.pyplot as plt
import seaborn as sns
from IPython.core.display import display, HTML
# --- plotly ---
from plotly import tools, subplots
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.express as px
import plotly.figure_factory as ff
import plotly.io as pio
pio.templates.default = "plotly_dark"
# --- models ---
from sklearn import preprocessing
from sklearn.model_selection import KFold
import lightgbm as lgb
import xgboost as xgb
import catboost as cb
# --- setup ---
pd.set_option('max_columns', 50)
載入資料
使用到的資料集: COVID-19/csse_covid_19_data/csse_covid_19_time_series/
- 全球
-
confirmed_global_df: dataframe,儲存從 2020/1/22 到現在的各國確診人數 -
deaths_global_df: dataframe,儲存從 2020/1/22 到現在各國死亡人數 -
recovered_global_df: dataframe,儲存從 2020/1/22 到現在各國康復人數
-
- 美國
-
confirmed_us_df: dataframe,儲存從 2020/1/22 到現在美國確診人數 -
deaths_us_df: dataframe,儲存從 2020/1/22 到現在美國死亡人數
-
import requests
for filename in ['time_series_covid19_confirmed_global.csv',
'time_series_covid19_deaths_global.csv',
'time_series_covid19_recovered_global.csv',
'time_series_covid19_confirmed_US.csv',
'time_series_covid19_deaths_US.csv']:
print(f'Downloading {filename}')
url = f'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/{filename}'
myfile = requests.get(url)
open(filename, 'wb').write(myfile.content)
資料前處理
變更日期的格式,由 mm/dd/yy 改成 yy-mm-dd
from datetime import datetime
def _convert_date_str(df):
try:
df.columns = list(df.columns[:4]) + [datetime.strptime(d, "%m/%d/%y").date().strftime("%Y-%m-%d") for d in df.columns[4:]]
except:
print('_convert_date_str failed with %y, try %Y')
df.columns = list(df.columns[:4]) + [datetime.strptime(d, "%m/%d/%Y").date().strftime("%Y-%m-%d") for d in df.columns[4:]]
confirmed_global_df = pd.read_csv('time_series_covid19_confirmed_global.csv')
_convert_date_str(confirmed_global_df)
deaths_global_df = pd.read_csv('time_series_covid19_deaths_global.csv')
_convert_date_str(deaths_global_df)
recovered_global_df = pd.read_csv('time_series_covid19_recovered_global.csv')
_convert_date_str(recovered_global_df)
將所有日期合併到同一欄位 (Date) ,該日期的累積人數合併到另一欄位 (Confirmed, Deaths, Recovered)
# Filter out problematic data points (The West Bank and Gaza had a negative value, cruise ships were associated with Canada, etc.)
removed_states = "Recovered|Grand Princess|Diamond Princess"
removed_countries = "US|The West Bank and Gaza"
confirmed_global_df.rename(columns={"Province/State": "Province_State", "Country/Region": "Country_Region"}, inplace=True)
deaths_global_df.rename(columns={"Province/State": "Province_State", "Country/Region": "Country_Region"}, inplace=True)
recovered_global_df.rename(columns={"Province/State": "Province_State", "Country/Region": "Country_Region"}, inplace=True)
confirmed_global_df = confirmed_global_df[~confirmed_global_df["Province_State"].replace(np.nan, "nan").str.match(removed_states)]
deaths_global_df = deaths_global_df[~deaths_global_df["Province_State"].replace(np.nan, "nan").str.match(removed_states)]
recovered_global_df = recovered_global_df[~recovered_global_df["Province_State"].replace(np.nan, "nan").str.match(removed_states)]
confirmed_global_df = confirmed_global_df[~confirmed_global_df["Country_Region"].replace(np.nan, "nan").str.match(removed_countries)]
deaths_global_df = deaths_global_df[~deaths_global_df["Country_Region"].replace(np.nan, "nan").str.match(removed_countries)]
recovered_global_df = recovered_global_df[~recovered_global_df["Country_Region"].replace(np.nan, "nan").str.match(removed_countries)]
將所有日期合併到同一欄位 (Date) ,該日期的累積人數合併到另一欄位 (Confirmed, Deaths, Recovered)
confirmed_global_melt_df = confirmed_global_df.melt(
id_vars=['Country_Region', 'Province_State', 'Lat', 'Long'], value_vars=confirmed_global_df.columns[4:], var_name='Date', value_name='ConfirmedCases')
deaths_global_melt_df = deaths_global_df.melt(
id_vars=['Country_Region', 'Province_State', 'Lat', 'Long'], value_vars=confirmed_global_df.columns[4:], var_name='Date', value_name='Deaths')
recovered_global_melt_df = deaths_global_df.melt(
id_vars=['Country_Region', 'Province_State', 'Lat', 'Long'], value_vars=confirmed_global_df.columns[4:], var_name='Date', value_name='Recovered')
recovered_global_melt_df.head()
製作訓練用的美國的資料集
train = confirmed_global_melt_df.merge(deaths_global_melt_df, on=['Country_Region', 'Province_State', 'Lat', 'Long', 'Date'])
train = train.merge(recovered_global_melt_df, on=['Country_Region', 'Province_State', 'Lat', 'Long', 'Date'])
# --- US ---
confirmed_us_df = pd.read_csv('time_series_covid19_confirmed_US.csv')
deaths_us_df = pd.read_csv('time_series_covid19_deaths_US.csv')
# 丟掉不須用到的欄位
confirmed_us_df.drop(['UID', 'iso2', 'iso3', 'code3', 'FIPS', 'Admin2', 'Combined_Key'], inplace=True, axis=1)
deaths_us_df.drop(['UID', 'iso2', 'iso3', 'code3', 'FIPS', 'Admin2', 'Combined_Key', 'Population'], inplace=True, axis=1)
# 將 Long_ 欄位改名為 Long
confirmed_us_df.rename({'Long_': 'Long'}, axis=1, inplace=True)
deaths_us_df.rename({'Long_': 'Long'}, axis=1, inplace=True)
# 變更日期的格式,由 mm/dd/yy 改成 yy-mm-dd
_convert_date_str(confirmed_us_df)
_convert_date_str(deaths_us_df)
# clean
# 丟掉不屬於美國的地區
confirmed_us_df = confirmed_us_df[~confirmed_us_df.Province_State.str.match("Diamond Princess|Grand Princess|Recovered|Northern Mariana Islands|American Samoa")]
deaths_us_df = deaths_us_df[~deaths_us_df.Province_State.str.match("Diamond Princess|Grand Princess|Recovered|Northern Mariana Islands|American Samoa")]
# --- Aggregate by province state ---
#confirmed_us_df.groupby(['Country_Region', 'Province_State'])
confirmed_us_df = confirmed_us_df.groupby(['Country_Region', 'Province_State']).sum().reset_index()
deaths_us_df = deaths_us_df.groupby(['Country_Region', 'Province_State']).sum().reset_index()
# remove lat, long.
confirmed_us_df.drop(['Lat', 'Long'], inplace=True, axis=1)
deaths_us_df.drop(['Lat', 'Long'], inplace=True, axis=1)
# 合併日期
confirmed_us_melt_df = confirmed_us_df.melt(
id_vars=['Country_Region', 'Province_State'], value_vars=confirmed_us_df.columns[2:], var_name='Date', value_name='ConfirmedCases')
deaths_us_melt_df = deaths_us_df.melt(
id_vars=['Country_Region', 'Province_State'], value_vars=deaths_us_df.columns[2:], var_name='Date', value_name='Deaths')
# 將美國確診及死亡的資料合併
train_us = confirmed_us_melt_df.merge(deaths_us_melt_df, on=['Country_Region', 'Province_State', 'Date'])
train = pd.concat([train, train_us], axis=0, sort=False)
train_us.rename({'Country_Region': 'country', 'Province_State': 'province', 'Date': 'date', 'ConfirmedCases': 'confirmed', 'Deaths': 'fatalities'}, axis=1, inplace=True)
train_us['country_province'] = train_us['country'].fillna('') + '/' + train_us['province'].fillna('')
train
train.rename({'Country_Region': 'country', 'Province_State': 'province', 'Id': 'id', 'Date': 'date', 'ConfirmedCases': 'confirmed', 'Deaths': 'fatalities', 'Recovered': 'recovered'}, axis=1, inplace=True)
train['country_province'] = train['country'].fillna('') + '/' + train['province'].fillna('')
# test.rename({'Country_Region': 'country', 'Province_State': 'province', 'Id': 'id', 'Date': 'date', 'ConfirmedCases': 'confirmed', 'Fatalities': 'fatalities'}, axis=1, inplace=True)
# test['country_province'] = test['country'].fillna('') + '/' + test['province'].fillna('')
綜觀全球疫情走勢
觀察 ww_df 的各個屬性
ww_df = train.groupby('date')[['confirmed', 'fatalities']].sum().reset_index()
ww_df['new_case'] = ww_df['confirmed'] - ww_df['confirmed'].shift(1)
ww_df['growth_factor'] = ww_df['new_case'] / ww_df['new_case'].shift(1)
ww_df.tail()
把 ww_df 的 confirmed fatalities new_case 三個屬性分別出來,放在 ww_melt_df
ww_melt_df = pd.melt(ww_df, id_vars=['date'], value_vars=['confirmed', 'fatalities', 'new_case'])
ww_melt_df
全球確診/死亡案例 (折線圖)
- 2020/04/02 確診人數突破 1M,且死亡人數為 52K
- 2020/07/28 確診人數突破 10M,且死亡人數為 500K
- 2020/08/13 確診人數突破 20M,且死亡人數為 750K
- 2020/11/08 確診人數突破 50M,且死亡人數為 1.25M
fig = px.line(ww_melt_df, x="date", y="value", color='variable',
title="Worldwide Confirmed/Death Cases Over Time")
fig.show()
全球確診/死亡案例 (折線圖) (取對數)
如果某段期間為斜直線,代表該段期間的案例為指數增長
比較 2020/3 初和 2020/3 底,確診案例成長率的上升速度略為增加
fig = px.line(ww_melt_df, x="date", y="value", color='variable',
title="Worldwide Confirmed/Death Cases Over Time (Log scale)",log_y=True)
fig.show()
全球死亡率 (折線圖)
可以明顯看到,死亡率在 2020 年 5 月達到高峰 (7%),隨後開始下降,可能是因為醫療能量慢慢提升,或是更多潛在患者確診?
ww_df['mortality'] = ww_df['fatalities'] / ww_df['confirmed']
fig = px.line(ww_df, x="date", y="mortality", title="Worldwide Mortality Rate Over Time")
fig.show()
計算全球新增確診案例成長因子 (折線圖)
公式: 每日新增確診案例 / 昨日新增確診案例
- 大於1: 確診案例增加
- 小於1: 確診案例減少
ww_df['mortality'] = ww_df['fatalities'] / ww_df['confirmed']
fig = px.line(ww_df, x="date", y="mortality", title="Worldwide Mortality Rate Over Time")
fig.show()
分析各國確診與死亡人數變化
宣告 country_df 條列各國確診與死亡人數,並查看 country_df 的各個欄位
country_df = train.groupby(['date', 'country'])[['confirmed', 'fatalities']].sum().reset_index()
country_df.tail()
列出 dataset 的所有國家 (注意,台灣的標籤為 Taiwan* )
countries = country_df['country'].unique()
print(f'{len(countries)} countries are in dataset:\n{countries}')
列出所有國家確診案例的數量級
target_date = country_df['date'].max()
print('Date: ', target_date)
for i in [1, 10, 100, 1000, 10000, 100000, 1000000, 10000000]:
n_countries = len(country_df.query('(date == @target_date) & confirmed > @i'))
print(f'{n_countries} countries have more than {i} confirmed cases')
以柱狀圖表示
sns.set_style('white')
ax = sns.distplot(np.log10(country_df.query('date == @target_date')['confirmed'] + 1))
ax.set_xlim([0, 6])
ax.set_xticks(np.arange(7))
dist_plot = ax.set_xticklabels(['10', '100', '1k', '10k', '100k', '1M', '10M'])
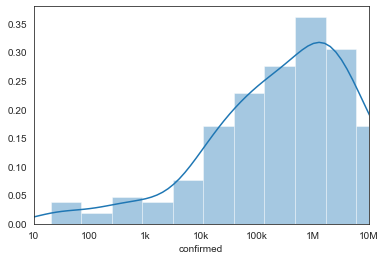
目前各國確診/死亡案例 (柱狀圖)
將各國的資料根據確診數排序,排除確診人數小於 1000 的國家 (此時台灣確診 911、 死亡 8),存在 top_country_df
下文確診案例、死亡案例、死亡率也一樣排除確診人數小於 1000 的國家
top_country_df = country_df.query('(date == @target_date) & (confirmed > 1000)').sort_values('confirmed', ascending=False)
top_country_melt_df = pd.melt(top_country_df, id_vars='country', value_vars=['confirmed', 'fatalities'])
fig = px.bar(top_country_melt_df.iloc[::-1],
x='value', y='country', color='variable', barmode='group',
title=f'Confirmed Cases/Deaths on {target_date}', text='value', height=1500, orientation='h')
fig.show()
目前確診案例前 10 多的國家 (折線圖)
top10_countries = top_country_df.sort_values('confirmed', ascending=False).iloc[:10]['country'].unique()
top10_countries_df = country_df[country_df['country'].isin(top10_countries)]
fig = px.line(top10_countries_df,
x='date', y='confirmed', color='country',
title=f'Confirmed Cases for top 10 country as of {target_date}')
fig.show()
目前死亡案例前 10 多的國家 (折線圖)
top10_countries = top_country_df.sort_values('fatalities', ascending=False).iloc[:10]['country'].unique()
top10_countries_df = country_df[country_df['country'].isin(top10_countries)]
fig = px.line(top10_countries_df,
x='date', y='fatalities', color='country',
title=f'Fatalities for top 10 country as of {target_date}')
fig.show()
目前死亡率最高的前 30 個國家 (柱狀圖)
top_country_df = country_df.query('(date == @target_date) & (confirmed > 100)')
top_country_df['mortality_rate'] = top_country_df['fatalities'] / top_country_df['confirmed']
top_country_df = top_country_df.sort_values('mortality_rate', ascending=False)
fig = px.bar(top_country_df[:30].iloc[::-1],x='mortality_rate', y='country',title=f'Mortality rate HIGH: top 30 countries on {target_date}', text='mortality_rate', height=800, orientation='h')
fig.show()
目前死亡率最低的前 30 個國家 (柱狀圖)
fig = px.bar(top_country_df[:-30].iloc[::-1], x='mortality_rate', y='country',title=f'Mortality rate LOW: top 30 countries on {target_date}', text='mortality_rate', height=800, orientation='h')
fig.show()
宣告 all_country_df 紀錄每個國家最近一天的疫情資訊,並新增以下欄位:
- 計算各國確診數的對數,記為
confirmed_log1p - 計算各國確診數的對數,記為
fatalities_log1p - 計算各國死亡率,記為
mortality_rate
all_country_df = country_df.query('date == @target_date')
all_country_df['confirmed_log1p'] = np.log10(all_country_df['confirmed'] + 1)
all_country_df['fatalities_log1p'] = np.log10(all_country_df['fatalities'] + 1)
all_country_df['mortality_rate'] = all_country_df['fatalities'] / all_country_df['confirmed']
all_country_df各國確診案例比較 (動態全球地圖) (以顏色區分等級)
顏色越深代表確診案例越多;反之則越少
fig = px.choropleth(all_country_df, locations="country",
locationmode='country names', color="confirmed_log1p",
hover_name="country", hover_data=["confirmed"],
range_color=[all_country_df['confirmed_log1p'].min(), all_country_df['confirmed_log1p'].max()],
color_continuous_scale="peach",
title='Countries with Confirmed Cases')
fig.show()各國死亡案例比較 (動態全球地圖) (以顏色區分等級)
顏色越深代表死亡數越多;反之則越少
fig = px.choropleth(all_country_df, locations="country",
locationmode='country names', color="fatalities_log1p",
hover_name="country", range_color=[0, 4],
hover_data=['fatalities'],
color_continuous_scale="peach",
title='Countries with fatalities')
fig.show()各國死亡率比較 (動態全球地圖) (以顏色區分等級)
顏色越深代表死亡率越高;反之則越低
fig = px.choropleth(all_country_df, locations="country",
locationmode='country names', color="mortality_rate",
hover_name="country", range_color=[0, 0.12],
color_continuous_scale="peach",
title='Countries with mortality rate')
fig.show()此時文中提到歐美澳的死亡率特別高,並提到一項假設是卡介苗在這些國家的接種率很高,也許跟卡介苗注射有關。不過考慮到這篇文章大概在 2020 年 5 到 6 月就有了,那時歐美的死亡率可能真的比較高,現在 (2021 年 2 月) 來看,歐美其實死亡率並沒有比較高。
死亡數演變 (折線圖) (取log) (超過 10 人死亡就開始記錄)
可以由這張圖比較各國的防疫能力 (醫療資源較充足、較早開始處理疫情的國家,曲線上升幅度會較緩慢)
n_countries = 10
n_start_death = 10
fatality_top_countires = top_country_df.sort_values('fatalities', ascending=False).iloc[:n_countries]['country'].values
country_df['date'] = pd.to_datetime(country_df['date'])
df_list = []
for country in fatality_top_countires:
this_country_df = country_df.query('country == @country')
start_date = this_country_df.query('fatalities > @n_start_death')['date'].min()
this_country_df = this_country_df.query('date >= @start_date')
this_country_df['date_since'] = this_country_df['date'] - start_date
this_country_df['fatalities_log1p'] = np.log10(this_country_df['fatalities'] + 1)
this_country_df['fatalities_log1p'] -= this_country_df['fatalities_log1p'].values[0]
df_list.append(this_country_df)
tmpdf = pd.concat(df_list)
tmpdf['date_since_days'] = tmpdf['date_since'] / pd.Timedelta('1 days')
fig = px.line(tmpdf, x='date_since_days', y='fatalities_log1p', color='country', title=f'Fatalities by country since 10 deaths, as of {target_date}')
fig.add_trace(go.Scatter(x=[0, 28], y=[0, 4], name='Double by 7 days', line=dict(dash='dash', color=('rgb(200, 200, 200)'))))
fig.add_trace(go.Scatter(x=[0, 56], y=[0, 4], name='Double by 14 days', line=dict(dash='dash', color=('rgb(200, 200, 200)'))))
fig.add_trace(go.Scatter(x=[0, 84], y=[0, 4], name='Double by 21 days', line=dict(dash='dash', color=('rgb(200, 200, 200)'))))
fig.show()
確診人數前十國每日新增確診案例 (折線圖)
country_df['prev_confirmed'] = country_df.groupby('country')['confirmed'].shift(1)
country_df['new_case'] = country_df['confirmed'] - country_df['prev_confirmed']
country_df['new_case'].fillna(0, inplace=True)
top10_country_df = country_df[country_df['country'].isin(top10_countries)]
fig = px.line(top10_country_df,
x='date', y='new_case', color='country',
title=f'DAILY NEW Confirmed cases by country')
fig.show()各國疫情發展情況: 確診案例 (點狀動態圖)
country_df['date'] = country_df['date'].apply(str)
country_df['confirmed_log1p'] = np.log1p(country_df['confirmed'])
country_df['fatalities_log1p'] = np.log1p(country_df['fatalities'])
fig = px.scatter_geo(country_df, locations="country", locationmode='country names',
color="confirmed", size='confirmed', hover_name="country",
hover_data=['confirmed', 'fatalities'],
range_color= [0, country_df['confirmed'].max()],
projection="natural earth", animation_frame="date",
title='COVID-19: Confirmed cases spread Over Time', color_continuous_scale="portland")
fig.show()由於所需的數據大於 512 KB ,超過 chart-stdio 的上限,這裡僅放截圖:

各國省分詳細資訊
提供詳細省分詳細資訊的僅有 8 個國家:
Australia, Canada, China, Denmark, France, Netherlands, US, UK
for country in countries:
province = train.query('country == @country')['province'].unique()
if len(province) > 1:
print(f'Country {country} has {len(province)} provinces: {province}\n')分析美國疫情
因為需要額外登入到 kaggle 下載資料,我就不搬運了,有興趣的可以看這裡
分析歐洲疫情
# Ref: https://www.kaggle.com/abhinand05/covid-19-digging-a-bit-deeper
europe_country_list =list([
'Austria','Belgium','Bulgaria','Croatia','Cyprus','Czechia','Denmark','Estonia','Finland','France','Germany','Greece','Hungary','Ireland',
'Italy', 'Latvia','Luxembourg','Lithuania','Malta','Norway','Netherlands','Poland','Portugal','Romania','Slovakia','Slovenia',
'Spain', 'Sweden', 'United Kingdom', 'Iceland', 'Russia', 'Switzerland', 'Serbia', 'Ukraine', 'Belarus',
'Albania', 'Bosnia and Herzegovina', 'Kosovo', 'Moldova', 'Montenegro', 'North Macedonia'])
country_df['date'] = pd.to_datetime(country_df['date'])
train_europe = country_df[country_df['country'].isin(europe_country_list)]
#train_europe['date_str'] = pd.to_datetime(train_europe['date'])
train_europe_latest = train_europe.query('date == @target_date')
fig = px.choropleth(train_europe_latest, locations="country",
locationmode='country names', color="confirmed",
hover_name="country", range_color=[1, train_europe_latest['confirmed'].max()],
color_continuous_scale='portland',
title=f'European Countries with Confirmed Cases as of {target_date}', scope='europe', height=800)
fig.show()歐洲國家確診案例 (折線圖)
train_europe_march = train_europe.query('date >= "2020-03-01"')
fig = px.line(train_europe_march, x='date', y='confirmed', color='country', title=f'Confirmed cases by country in Europe, as of {target_date}')
fig.show()歐洲國家死亡案例 (折線圖)
fig = px.line(train_europe_march, x='date', y='fatalities', color='country', title=f'Fatalities by country in Europe, as of {target_date}')
fig.show()歐洲每日新增確診案例 (折線圖)
train_europe_march['prev_confirmed'] = train_europe_march.groupby('country')['confirmed'].shift(1)
train_europe_march['new_case'] = train_europe_march['confirmed'] - train_europe_march['prev_confirmed']
fig = px.line(train_europe_march, x='date', y='new_case', color='country', title=f'DAILY NEW Confirmed cases by country in Europe')
fig.show()分析亞洲疫情
country_df['date'] = pd.to_datetime(country_df['date'])
country_latest = country_df.query('date == @target_date')
fig = px.choropleth(country_latest, locations="country", locationmode='country names', color="confirmed", hover_name="country", range_color=[1, 1000000],
color_continuous_scale='portland', title=f'Asian Countries with Confirmed Cases as of {target_date}', scope='asia', height=800)
fig.show()亞洲國家每日新增確診案例 (折線圖)
top_asian_country_df = country_df[country_df['country'].isin(['China', 'Indonesia', 'Iran', 'Japan', 'Korea, South', 'Malaysia', 'Philippines','India', 'Bangladesh', 'Pakistan', 'Saudi Arabia', 'Turkey'])]
fig = px.line(top_asian_country_df, x='date', y='new_case', color='country', title=f'DAILY NEW Confirmed cases Asia')
fig.update_layout(yaxis_range=[0, 100000])
fig.show()
哪些國家已經脫離疫情高峰
以下地圖顯示,黃色國家的比例很高,代表確診人數快速上升,處於疫情高峰;藍色和紫色國家的比率較低,代表從高峰期開始下降。
我們可以發現歐美似乎有漸漸脫離高峰期,東南亞正在疫情高峰,德國及澳洲似乎已將疫情控制
max_confirmed = country_df.groupby('country')['new_case'].max().reset_index()
country_latest = pd.merge(country_latest, max_confirmed.rename({'new_case': 'max_new_case'}, axis=1))
country_latest['new_case_peak_to_now_ratio'] = country_latest['new_case'] / country_latest['max_new_case']
recovering_country = country_latest.query('new_case_peak_to_now_ratio < 0.5')
major_recovering_country = recovering_country.query('confirmed > 100')
fig = px.choropleth(country_latest, locations="country", locationmode='country names', color="new_case_peak_to_now_ratio", hover_name="country", range_color=[0, 1], hover_data=['confirmed', 'fatalities', 'new_case', 'max_new_case'], title='Countries with new_case_peak_to_now_ratio')
fig.show()全球疫情何時趨緩
利用 sigmoid 函數建立一個模型協助我們預測各國的疫情何時趨緩 (確診人數不再暴增)
待續…